 PDF(3166 KB)
PDF(3166 KB)


CRISPR/Cas9 Technology: Exploring The Functions of lncRNAs and Their Roles in Cancer Progression
LI Xin, HU Ying, WANG Yu-Ming
Chinese Journal of Biochemistry and Molecular Biology ›› 2025, Vol. 41 ›› Issue (3) : 364-375.
 PDF(3166 KB)
PDF(3166 KB)
 PDF(3166 KB)
PDF(3166 KB)
CRISPR/Cas9 Technology: Exploring The Functions of lncRNAs and Their Roles in Cancer Progression
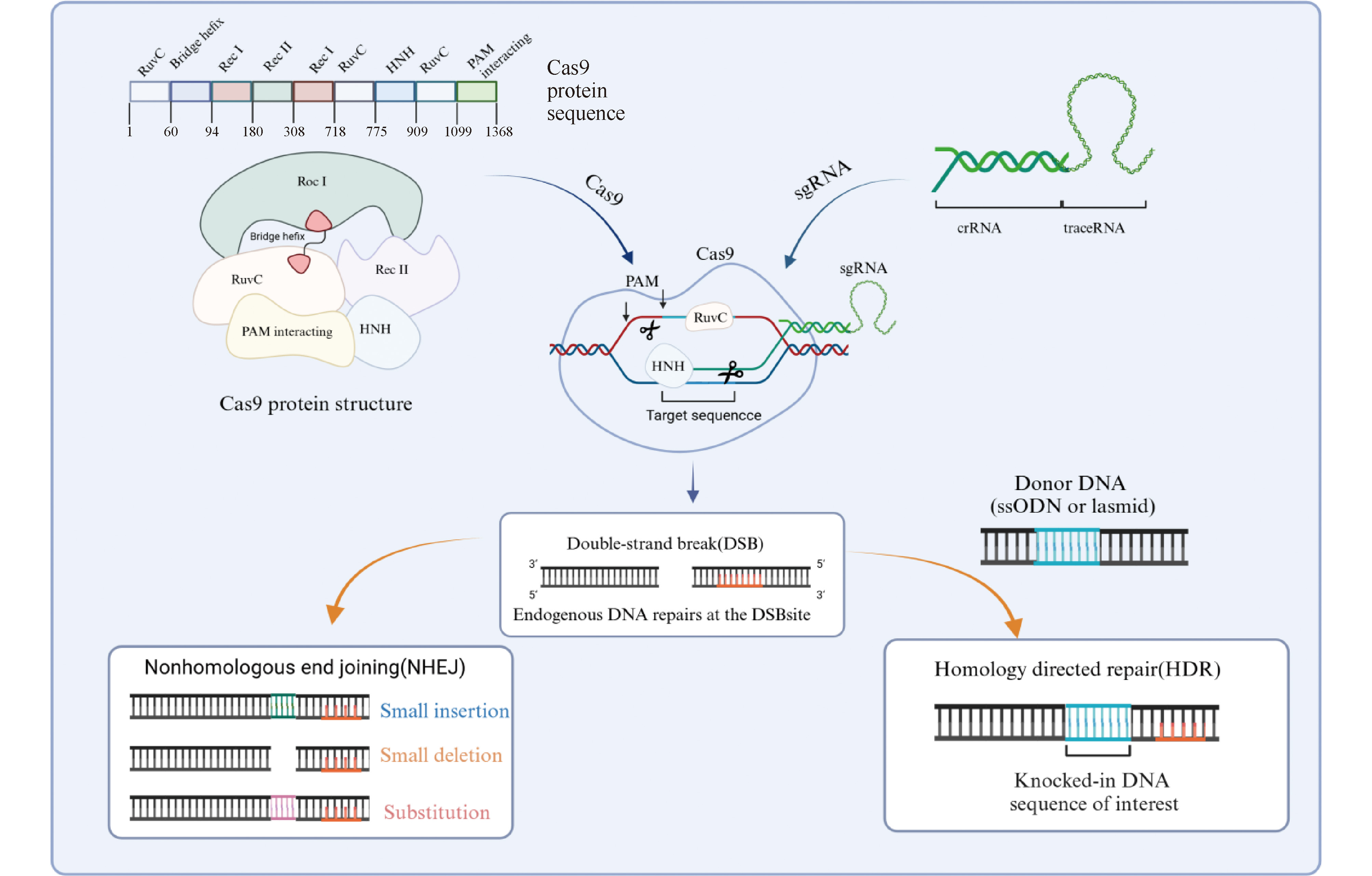
clustered regularly interspaced short palindromic repeats and CRISPR-related protein 9 (CRIAPR/Cas9) / long non-coding RNA (lncRNA) / gene regulation / cancer {{custom_keyword}} /
 PDF(3166 KB)
PDF(3166 KB)
/
| 〈 |
|
〉 |